New method paves the way for new antibiotics
NTNU has developed a promising antibiotic candidate against MRSA. Behind the discovery lies a methodology that may be important in the fight against antimicrobial resistance.
“Antimicrobial resistance is a major problem, and being able to help solve it is really great,” says Amanda Holstad Singleton, a PhD candidate.
It’s one thing to develop new antibiotic candidates which when combined prove to be well tolerated by human cells, but developing a technology to study how the antibiotic works inside the bacterial cells is equally important.
She is the lead author of a study that shows how a combination of two new substances effectively kills methicillin-resistant Staphylococcus aureus (MRSA).
The substances have been developed at NTNU and may become a completely new antibiotic that is effective against a wide group of bacteria.
“It’s one thing to develop new antibiotic candidates which when combined prove to be well tolerated by human cells, but developing a technology to study how the antibiotic works inside the bacterial cells is equally important,” says Singleton.
Red light for internal processes
To analyse how the two substances worked, the NTNU research team has developed a method that analyses how the bacterium’s signalling proteins react to the treatment. The method provides researchers with a completely new tool in the search for new antibiotic candidates.
“Up to 10,000 proteins can be found inside a bacterial cell. Instead of looking at all of them, we ‘fish’ out the 2000 or so proteins that are signalling proteins. These proteins control much of what happens in the cells,” says Singleton.
The method allows researchers to see whether each of these proteins is activated or deactivated after adding the substance they want to test.
“These proteins can be compared to traffic lights, which can change from red to green and back again. By getting them to change to red, you stop an important signalling pathway inside the cell,” says Singleton.
Amanda Holstad Singleton is the lead author in a study showing how the combination of two new substances effectively kills MRSA. The substances are well tolerated by human cells, making them a promising candidate for a new antibiotic. Photo: Jana Scheffold
If a substance is found to influence a signalling protein by switching the traffic light to red for a key process inside a cell, it is considered a candidate for a new antibiotic. If a substance is found to yield a red light for several different processes in a cell, it is an even better candidate.
That’s precisely what NTNU researchers have done after combining two different substances that could become a new antibiotic.
“In a study recently published in the Frontiers in Microbiology journal, we show that a combination of two new substances developed at NTNU kill MRSA much more effectively than when used separately,” says Singleton.
- You might also like: How children are affected by the coronavirus, RS virus and rhinoviruses
Prevents DNA copying
Approximately four years ago, researchers at NTNU’s Department of Clinical and Molecular Medicine published the bactericidal properties of a particular type of peptide. These peptides, in combination with a compound developed at NTNU’s Department of Chemistry, may now become a completely new type of antibiotic.
“Peptides are chains of amino acids, which are the building blocks of proteins. What is special about these particular peptides is that they bind to a protein in the bacteria that is absolutely essential for bacteria to be able to copy their DNA,” says Professor Marit Otterlei.
The peptide prevents DNA copying, and thus, the bacterium dies.
“No other antibiotics attack this protein. That means the protein is a new target, and consequently no bacteria are resistant to the peptides. Since this target protein is found in all bacteria, these peptides also work on multidrug-resistant bacteria,” says Otterlei.
Synergy effect
While Otterlei and her colleagues continued working on the peptides, researchers Eirik Sundby and Bård Helge Hoff at NTNU’s Department of Materials Science and Engineering and the Department of Chemistry were working to find substances that effectively prevented the formation of DNA building blocks. They had also developed compounds, known as kinase inhibitors, that could be used in the method of fishing out signalling proteins from the bacterial samples.
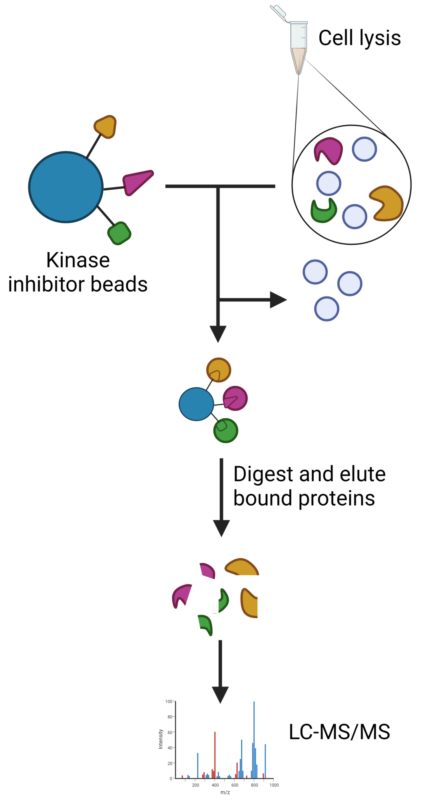
For people interested in the science behind these findings: By comparing cell lysis from untreated and treated bacteria, it is possible to see which proteins have changed. The cell lysis contains ‘traffic light proteins’, i.e. proteins involved in various bacterial signalling pathways. By using beads coupled to kinase inhibitors, activated proteins can be selected for via an available ATP-binding site (green traffic light). This provides insight into which cellular processes are active or deactivated, and thus provides valuable information about a cell’s response to antibiotics. Ill: Amanda Holstad Singleton
“When the method was ready, we tested it on bacteria treated with peptides in combination with one of these new molecules that was thought to affect the production of DNA building blocks. We found that the new molecules had a different mechanism of action than we thought, but they did produce a very good combination effect with our peptides, a so-called synergistic effect,” says Otterlei.
It turned out that the new molecules developed by Sundby and Hoff inhibited energy metabolism inside the bacterial cell. In combination with Otterlei’s peptides, they also resulted in the activation of proteins linked to multiple stress responses in the bacterial cells. This did not happen when the substances were administered separately. This extra activation caused the bacteria to die more efficiently.
This is the first time the effectiveness of antibiotics has been studied in this way, the researchers say.
“This gives us a completely new way of assessing new antibiotic candidates,” says Otterlei.
- You might also like: Why you should be vaccinated even if you have had COVID-19
Prevents mutations that can cause resistance
It also provides researchers with a new way to prevent the development of resistance to new antibiotics.
“We must remember that developing resistance is a natural part of evolution. It is inevitable. However, developing resistance is costly for the bacterium. It has to make some sacrifices,” says Otterlei.
Singleton explains that there are two ways bacteria can develop resistance to antibiotics: either by the bacterium coming into contact with other bacteria that are already resistant and exchanging DNA among themselves, or that there is a mutation in the bacterium’s genes that protects it against the antibiotic.
“This type of mutation comes at a cost, it affects the bacteria’s fitness. One trait is sacrificed in order to obtain another that provides protection against the antibiotic.
If the advantage of being protected against the antibiotic outweighs the disadvantage, the bacterium will multiply, and we get many new antibiotic-resistant bacteria.
However, if the bacterium has to develop resistance to two substances at the same time, which work in completely different places inside the bacterial cell, the job becomes a lot harder.
If you attack two different processes, developing resistance to both will be too much of a burden, and the bacteria will become less viable,” says Singleton.
It becomes even more difficult if you also create an antibiotic that attacks the very way the bacterium develops resistance.
“In our case, the protein that our new antibiotic candidate attacks plays such a key role in copying the bacterium’s DNA before it can divide, that if a mutation occurs, the loss of fitness becomes so great that the bacterium dies,” says Singleton.
Source:
Amanda Holstad Singleton, Marit Ottelei et al: Frontiers | Activation of multiple stress responses in Staphylococcus aureus substantially lowers the minimal inhibitory concentration when combining two novel antibiotic drug candidates Front. Microbiol., 25 September 2023. Sec. Antimicrobials, Resistance and Chemotherapy
Volume 14 – 2023 | https://doi.org/10.3389/fmicb.2023.1260120